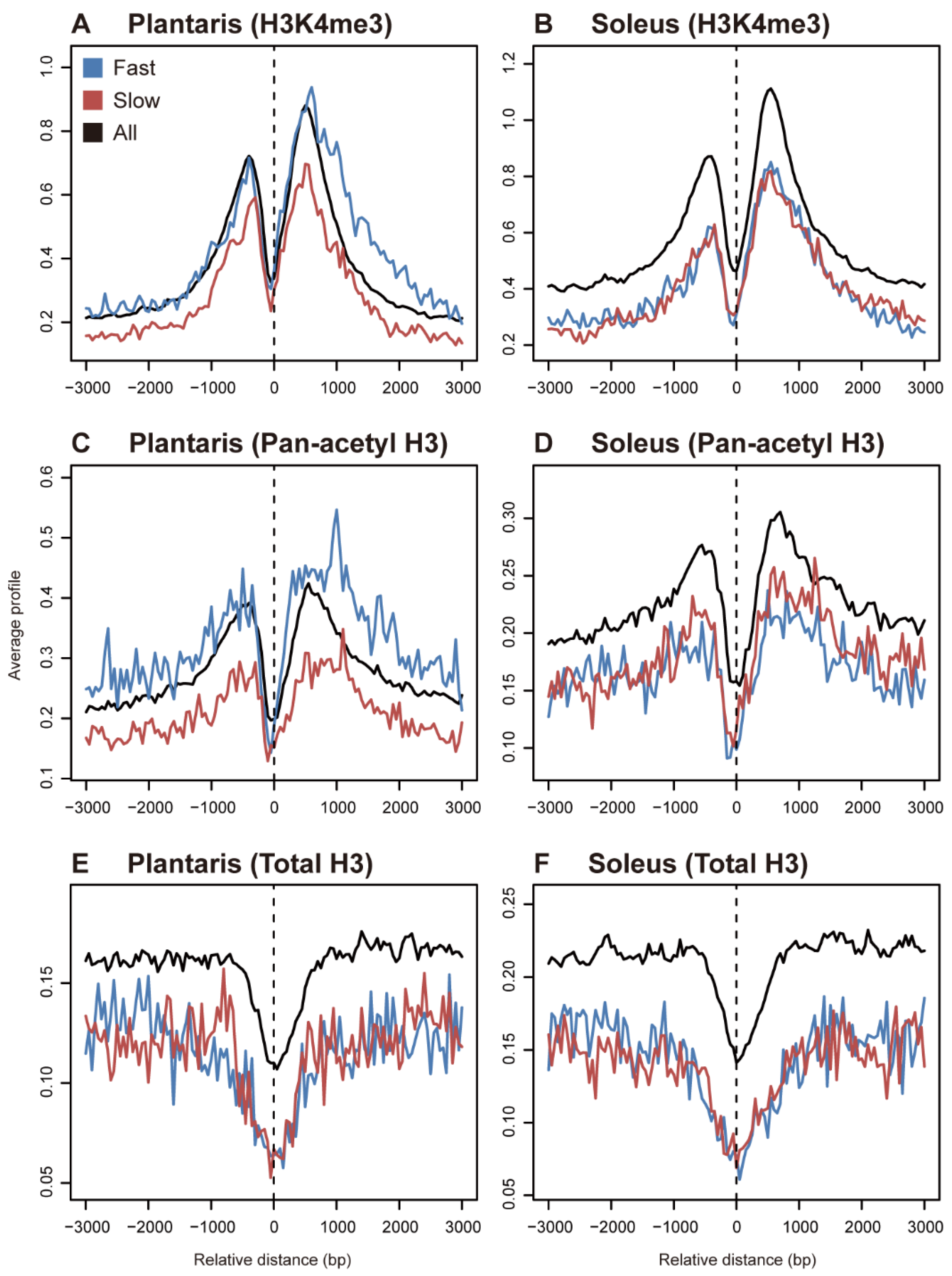
wide analysis of histone modification patterns in Biology Diagrams Accumulation of reverse centromeric transcripts is still cell-cycle dependent in cells lacking Dicer, consistent with an involvement of the exosome at the end of S phase . The exosome affects the turnover of other S-phase-specific transcripts, such as histone transcripts in Saccharomyces cerevisiae , perhaps by a similar mechanism [11,12] . The histone modifications are lowest at the LS cell-cycle phase (Figure 2E, late on bottom), consistent with chromatin mark dilution during replication. Genomic regions in the blue H3K27me3-enriched state ( Figure 2 F, histone modification heatmap) have a wide range of temporal changes coordinated between histone methylation marks, chromatin Here we use cell cycle sorting and ChIP-seq to analyse the genomic distribution of three histone modifications closely associated with transcriptional regulation (H3K9ac, H3K4me3 and H3K27me3

López-Hernández et al. review how transcription, replication, and repair shape complex patterns of histone modifications on chromatin, processes that must be collectively interpreted for proper genome function. In this model, histone modifications initiate, maintain, and record interactions between different DNA processes, facilitating communication between them. Epigenetic states defined by chromatin can be maintained through mitotic cell division. Alabert et al. tracked histone modifications and histone variants during DNA replication and across the cell cycle and found that post-translational modifications (PTMs) are transmitted with parental histones to newly replicated DNA. It is thus often challenging to quantitatively compare histone modification ChIP-seq data sets from different phases of the cell cycle. To overcome this, we used a spike-in based normalization strategy, whereby adding an equal amount of exogenous chromatin from a different species into each sample can be used as a more accurate normalization
Regulation of chromatin by histone modifications Biology Diagrams
In eukaryotic cells, posttranslational modifications (PTMs) on histones regulate chromatin structure and thus impact nearly all chromatin-templated events, including replication, transcription, and DNA repair. This chapter describes strategies for investigating the reestablishment of histone PTMs during the mitotic cell cycle using

Histone lysine acetylation (Kac) is a classical histone acylation that has an effect related to DNA damage and repair, cell-cycle control, chromatin architecture, RNA splicing, and transcription . Methylation takes part in gene repression or activation, depending on the sites and degree [ 14 , 15 ]. For example, repression of the cell-cycle-dependent cyclin E gene by the retinoblastoma gene product RB involves recruitment of HDACs, Here, we describe the known histone modifications, define We highlight recent advances in understanding cell cycle-dependent histone biogenesis and histone modification deposition, how cell cycle regulators control histone modifier activities, the contribution of chromatin modifications to origin firing for DNA replication, and newly identified roles for nucleoporins in regulating cell cycle gene